New version of SpecTcl (1st implemented in May 2023)
Instructions to get started
In this new version of SpecTcl, written by Giordano Cerizza and Simon Giraud in Python, Xamine is replaced for online and offline data analysis by the CutiePie window, while the TreeGui and Spectcl control windows are retained from the previous version. To use this version, follow these instructions:
- While in the S800 account, issue from any directory goQtspectcl_s800. This command will open the TreeGui, the SpecTcl control window and log window, and the CutiePie window, which are all shown below. If it happens that Xamine is still opened instead of QtPy, close this SpecTcl session, and remove the comment on the line “set DisplayType qtpy” of “/user/s800/s800develop/s800Qtspectcl/SpecTclInit.tcl”, before issuing the command to start SpecTcl with QtPy again.
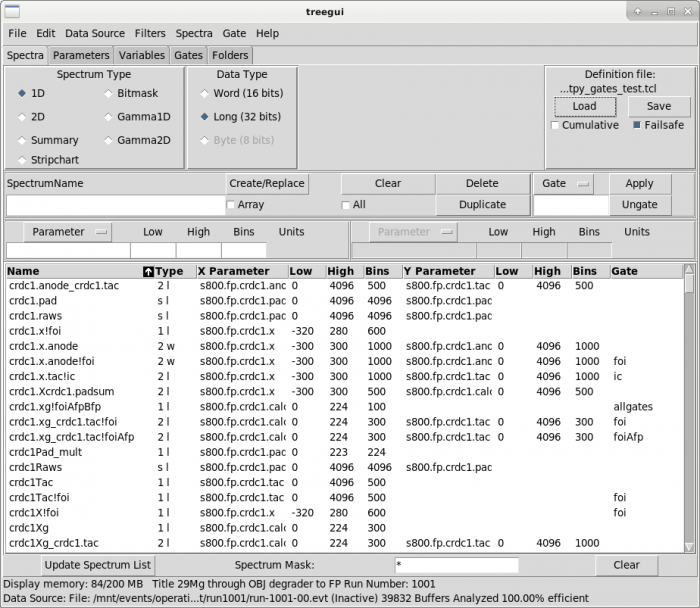
- In the TreeGui, load the definition file.
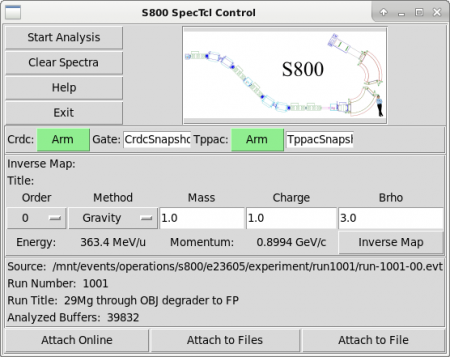
- In the SpecTcl control window, click 'Attach to file' in the bottom-right, to attach to data file (of type .evt) and let it run the data.
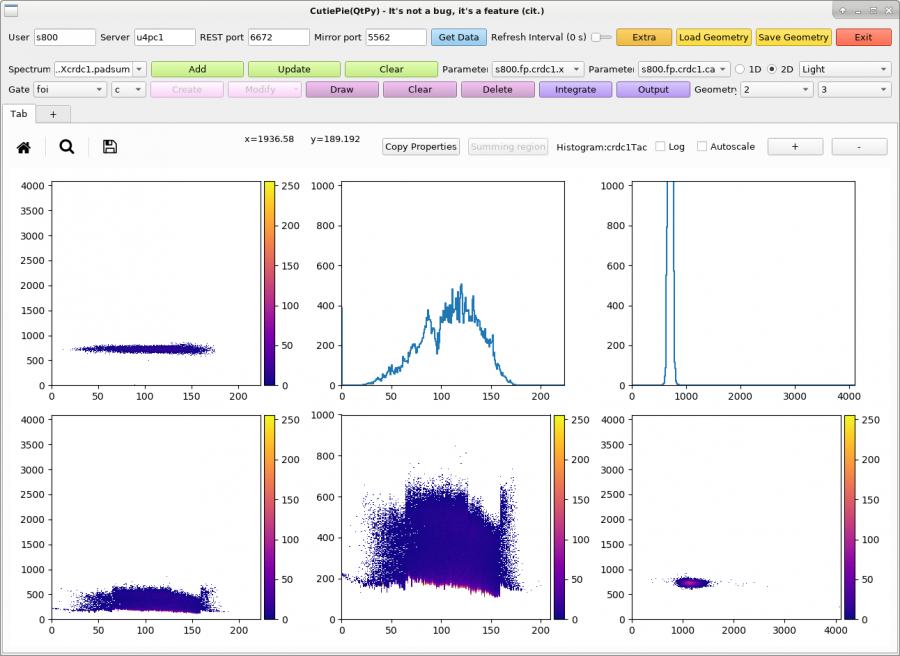
- Now click the light-blue button called 'Get Data' on CutiePie.
- On CutiePie, click 'Load Geometry' and select the folder 'Windows/qtpy'. Here you will open a file of type *qtpy.win which will open a set of spectra from the data you want to look at.
You can now see and inspect your data set using CutiePie.
The CutiePie window with geometry file for CRDC1 spectra. The data are from Run 1001 of Experiment 23605 in 2023, which utilized a 29Mg beam.
Useful tips and tricks
- You can see the title of a spectrum by sliding your mouse over it.
- At the top of the CutiePie window you will see two rows of buttons, one for functionalities related to the selected spectrum and one below it related to the gates.
- You can modify the geometry using two buttons in the top-right of the CutiePie window where you can adjust the numbers of rows and columns.
- To make a gate on a given spectrum, double-click on the spectrum, so the 'Create button becomes clickable, then click on the 'Create' button. You can then choose a name for your gate and thereafter also the type e.g. 'c' for contour. You can left-click on different points of the spectrum to draw lines for this contour gate. Double-click to end and save the gate. This gate should now also show up in the TreeGui.
- Double-click to work on only one spectrum. Double-click again to return to multiple panels.
- After selecting a gate, you can integrate over it using the purple 'Integrate' button.
- With the 'summing' button you can select the regions to sum over by using left-click twice, and then double-click to close the summing region (for 1-d case). In 2-d you will make multiple left-clicks and again double-click to close the summing region.
- With the magnifying glass, you can zoom into a spectrum using click and drag. Click again on the magnifying glass to set the zoom. Click on the Home button to reset it.
- You can also use the 'Copy properties' button to implement the gates from one spectrum onto another. This can only be used between spectra of the same dimensions.
- To make a fit, click on the 'Extra' button, which will open a new window with additional functionalities. First click on the spectrum you wish to fit, before selecting the type and region of fit.
- Also from the 'Extra' window, you can overlay images from anywhere on your computer using the 'Overlay' button. The overlayed image can be moved around, contracted or expanded using the 'joystick' control with the black circle at its center.
If you detect any bugs or have any questions not addressed here, please send a message to Simon Giraud at Giraud@frib.msu.edu. For more information on CutiePie, refer to this page: https://docs.nscl.msu.edu/daq/newsite/qtpy/index.html, which can also be accessed through the NSCL DAQ page: https://docs.nscl.msu.edu/daq/newsite/index.php.